New Nanopore Technology Enables Amplification-Free Quantification of Endogenous Mitochondrial DNA, Promising Advances in Disease Diagnostics
A team led by the postdoctoral fellow Dr. Sohini Pal and headed by Prof. Amit Meller from the Technion Faculty of Biomedical Engineering developed a novel method using solid-state nanopores and machine learning to directly identify and count mitochondrial DNA molecules without the need for amplification, offering a potentially more accurate and accessible tool for studying diseases linked to mitochondrial dysfunction, such as cancer and neurodegenerative disorders.
Mitochondria, the powerhouses of cells, contain their own DNA (mtDNA). The number of mtDNA copies can vary significantly and changes are associated with aging, altered energy balance, and various diseases. Traditionally, quantifying mtDNA relies on methods like qPCR, which require amplifying small segments of DNA. However, these amplification steps can introduce biases and inaccuracies, limiting their reliability for precise clinical diagnostics.
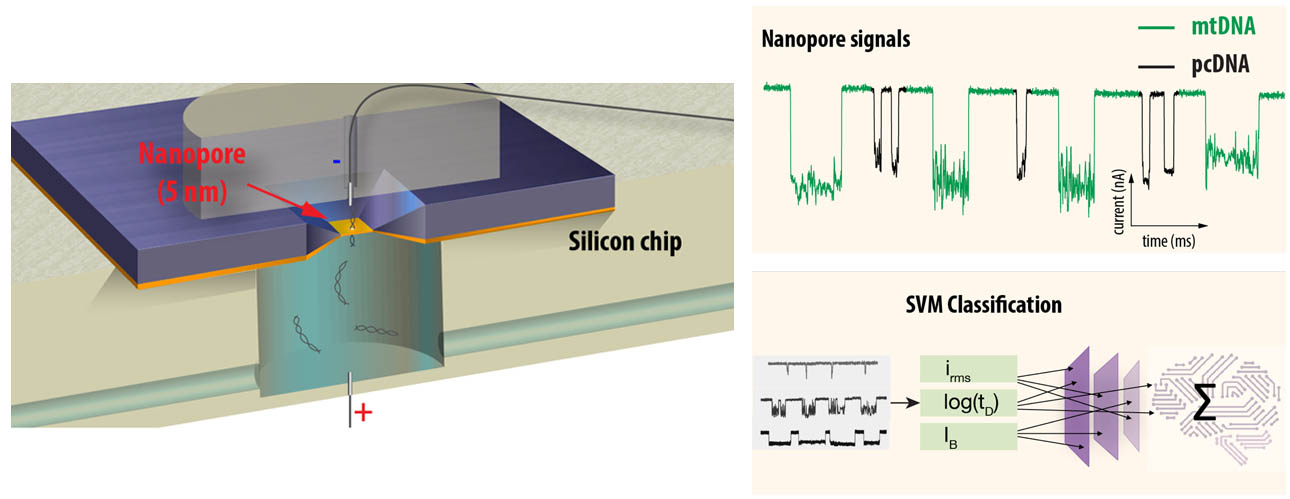
This new study introduces an amplification-free approach leveraging solid-state nanopore (ssNP) technology developed at the Technion. Nanopores are tiny holes through which individual DNA molecules can be passed, generating unique electrical signals (ion-current signatures) as they traverse the pore. These signals contain rich information about the molecule's length and structure.
Intriguingly, the research team discovered that native mtDNA molecules, extracted directly from biological samples, produce distinct electrical signatures compared to synthetic DNA or linearized plasmid DNA of similar lengths. These unique signatures, characterized by specific current fluctuations, appear to stem from the mtDNA's native structure, potentially including associated proteins which are typically lost during amplification-based sample preparation. A machine learning model developed by the team identifies the mtDNA molecules one by one, distinguishing them from other forms of DNA such as genomic DNA, that are present in the sample.
This innovative approach represents a significant step forward in mtDNA analysis, offering a sensitive, reliable, and efficient method for quantifying mtDNA copy numbers directly from biological samples. Its potential for rapid diagnostics in clinical settings could improve the understanding and management of mitochondrial-related diseases.
The study was published in ACS Nano on March 13, 2025.